outline¶
- R: what is it for ?
- R: base syntax
- R: the packages you need to know/learn to be efficient
- Demos on physicists data
- The pitfalls to avoid
- how to learn and where to look
R is created in 1995
An open source implementation of S (Bell labs statistical software)
Designed to make data analysis and statistics easy
interpreted language, high level
Primarily used in academics, moving to enterprise lately
Pros¶
- open source, so free
- Wonderful IDE: Rstudio
- variable exploration
- help, plot tools
- github integrated
- huge community! so lot of helps
- Lots of tools (libraries)
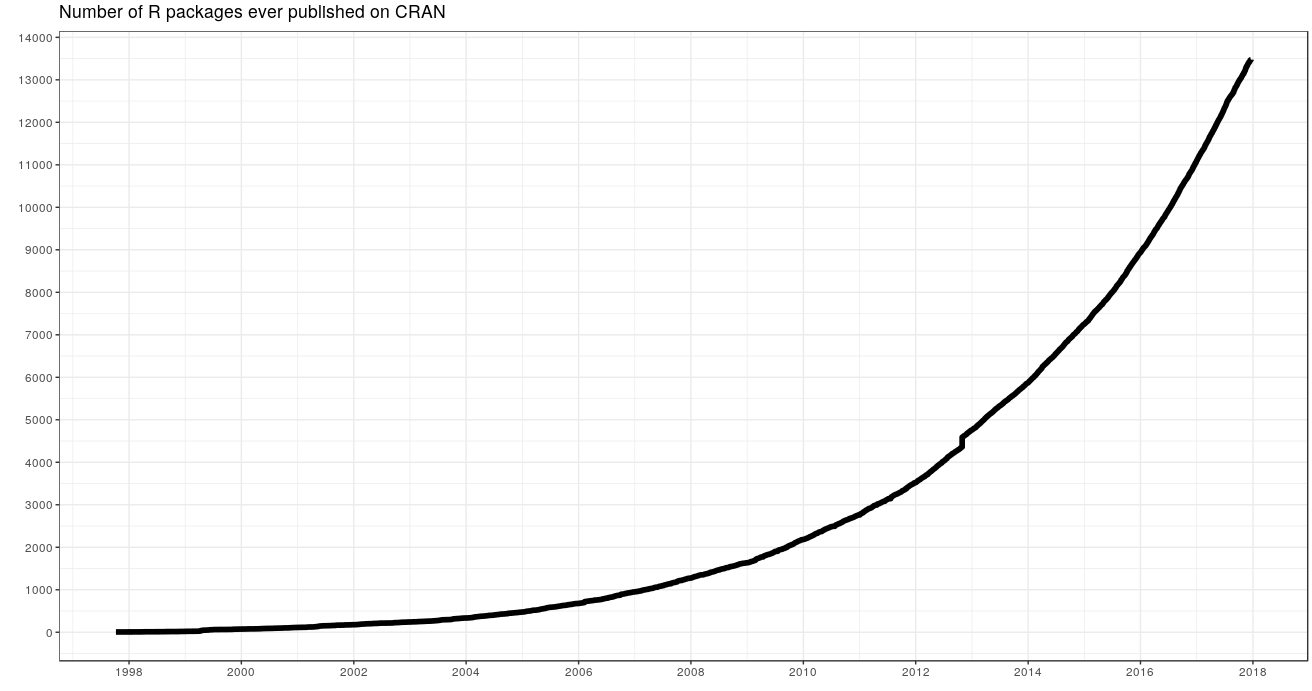
- easy to paralelize, can be used on baobab
Cons¶
- Slow (not for Whab or Nico, but some library are coded in C)
- lots of ways to do the same things, so a bit messy in the beginning
- hard to find the appropriates tools in the beginning (but i ll give you the good ones)
- steep learning curve, but accesible for experienced programmers or physicists
Basic syntax¶
When you install R, it comes with base package and 6 others.
R is interpreted, and guess the type you declare (like matlab and python)
a <- 2
b = 2
a
b
data type¶
a <- "plouf"
class(a)
b <- 2
class(b)
c <- 2L
class(c)
d <- as.factor("plouf")
class(d)
WTF factor ?
a
d
as.numeric(a)
as.numeric(d)
Warning message in eval(expr, envir, enclos): "NAs introduits lors de la conversion automatique"
used in statistical model (generalised linear models, etc), but dangerous
data structure type¶
vector¶
# vector
a <- c(2,5,7,8)
a
class(a)
- 2
- 5
- 7
- 8
a[1]
a[1:2]
- 2
- 5
a[1,4]
Error in a[1, 4]: nombre de dimensions incorrect Traceback:
a[c(1,4)]
- 2
- 8
everything same class, so coerce things
a <- c(2,5,7,"plouf")
class(a)
a[1]
matrix¶
b <- matrix(c(3,4,5,8),nrow = 2, ncol = 2)
b
| 3 | 5 |
| 4 | 8 |
b[1,]
- 3
- 5
b[,2]
- 5
- 8
b[1]
all matrix operation exists, equation solving etc.
list¶
c <- list(trucs = c(1,5,7), plouf = "des choses", matrice = matrix(sample(1:100,25),5,5) )
c
- $trucs
- 1
- 5
- 7
- $plouf
- 'des choses'
- $matrice
63 31 28 78 29 44 57 46 38 34 76 84 95 51 83 17 15 36 25 66 12 81 100 69 70
c$trucs
c[[1]]
c[["trucs"]]
c["trucs"]
- 1
- 5
- 7
- 1
- 5
- 7
- 1
- 5
- 7
- 1
- 5
- 7
data frame: une liste avec contrainte de dimension !¶
C'est le coeur du traitement de données dans R
d <- data.frame(experiment = rep(LETTERS[1:5], each = 2), data1 = sample(c(0,1),10,replace = T), data2 = sample(1:100,10))
d
| experiment | data1 | data2 |
|---|---|---|
| A | 1 | 98 |
| A | 1 | 22 |
| B | 0 | 87 |
| B | 1 | 85 |
| C | 1 | 52 |
| C | 1 | 72 |
| D | 0 | 84 |
| D | 1 | 38 |
| E | 0 | 75 |
| E | 1 | 16 |
#idem list:
d$experiment
d[,"experiment"]
- A
- A
- B
- B
- C
- C
- D
- D
- E
- E
- A
- A
- B
- B
- C
- C
- D
- D
- E
- E
d$experiment[1]
d[1,"experiment"]
d[1,1]
Esay to subset using boolean.
d$experiment
d$experiment == "A"
- A
- A
- B
- B
- C
- C
- D
- D
- E
- E
- TRUE
- TRUE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
- FALSE
d[d$experiment == "A",]
| experiment | data1 | data2 |
|---|---|---|
| A | 1 | 98 |
| A | 1 | 22 |
loops and conditional¶
d[d$data2 > 50 & d$data1 == 0,]
| experiment | data1 | data2 | |
|---|---|---|---|
| 3 | B | 0 | 87 |
| 7 | D | 0 | 84 |
| 9 | E | 0 | 75 |
d[d$data2 > 50 | d$data1 == 0,]
| experiment | data1 | data2 | |
|---|---|---|---|
| 1 | A | 1 | 98 |
| 3 | B | 0 | 87 |
| 4 | B | 1 | 85 |
| 5 | C | 1 | 52 |
| 6 | C | 1 | 72 |
| 7 | D | 0 | 84 |
| 9 | E | 0 | 75 |
for(i in 1:4){
if(i>2){
print(paste0(i," est sup à 2" ))}else{
print(paste0(i," est inf à 2" ))}
}
[1] "1 est inf à 2" [1] "2 est inf à 2" [1] "3 est sup à 2" [1] "4 est sup à 2"
Specific of R, really usefull: lapply and sapply (and others)
plouf <- lapply(1:4,function(x){x^2})
plouf
class(plouf)
plouf2 <- sapply(1:4,function(x){x^2})
plouf2
class(plouf2)
- 1
- 4
- 9
- 16
- 1
- 4
- 9
- 16
sapply(c("data1","data2"),function(col){
summary(d[,col])
})
| data1 | data2 | |
|---|---|---|
| Min. | 0.0 | 5.00 |
| 1st Qu. | 0.0 | 21.25 |
| Median | 0.0 | 47.50 |
| Mean | 0.4 | 46.60 |
| 3rd Qu. | 1.0 | 59.25 |
| Max. | 1.0 | 95.00 |
Libraries¶
library(ggplot2) # pour les graphiques
head(mpg)
| manufacturer | model | displ | year | cyl | trans | drv | cty | hwy | fl | class |
|---|---|---|---|---|---|---|---|---|---|---|
| audi | a4 | 1.8 | 1999 | 4 | auto(l5) | f | 18 | 29 | p | compact |
| audi | a4 | 1.8 | 1999 | 4 | manual(m5) | f | 21 | 29 | p | compact |
| audi | a4 | 2.0 | 2008 | 4 | manual(m6) | f | 20 | 31 | p | compact |
| audi | a4 | 2.0 | 2008 | 4 | auto(av) | f | 21 | 30 | p | compact |
| audi | a4 | 2.8 | 1999 | 6 | auto(l5) | f | 16 | 26 | p | compact |
| audi | a4 | 2.8 | 1999 | 6 | manual(m5) | f | 18 | 26 | p | compact |
ggplot(data = mpg) +
geom_point(aes(x = displ, y = hwy, color = class))
ggplot(data = mpg) +
geom_point(aes(x = displ, y = hwy)) +
facet_grid(drv ~ cyl)
library(data.table) # pour le data management
dt <- setDT(copy(mpg))
class(dt)
- 'data.table'
- 'data.frame'
plus <- data.table(manufacturer = c('audi','chevrolet','dodge','ford','honda','hyundai','jeep','land rover','lincoln','mercury','nissan','pontiac','subaru','toyota','volkswagen'),
country = c("DE","US","US","US","JP","JP","US","UK","UK","US","JP","US","DE","JP","DE"))
head(plus)
| manufacturer | country |
|---|---|
| audi | DE |
| chevrolet | US |
| dodge | US |
| ford | US |
| honda | JP |
| hyundai | JP |
merged <- merge(dt,plus,on = "manufacturer")
head(merged)
| manufacturer | model | displ | year | cyl | trans | drv | cty | hwy | fl | class | country |
|---|---|---|---|---|---|---|---|---|---|---|---|
| audi | a4 | 1.8 | 1999 | 4 | auto(l5) | f | 18 | 29 | p | compact | DE |
| audi | a4 | 1.8 | 1999 | 4 | manual(m5) | f | 21 | 29 | p | compact | DE |
| audi | a4 | 2.0 | 2008 | 4 | manual(m6) | f | 20 | 31 | p | compact | DE |
| audi | a4 | 2.0 | 2008 | 4 | auto(av) | f | 21 | 30 | p | compact | DE |
| audi | a4 | 2.8 | 1999 | 6 | auto(l5) | f | 16 | 26 | p | compact | DE |
| audi | a4 | 2.8 | 1999 | 6 | manual(m5) | f | 18 | 26 | p | compact | DE |
merged[country == "DE",.(mean_comsum = mean(hwy)),by = .(manufacturer,model)]
| manufacturer | model | mean_comsum |
|---|---|---|
| audi | a4 | 28.28571 |
| audi | a4 quattro | 25.75000 |
| audi | a6 quattro | 24.00000 |
| subaru | forester awd | 25.00000 |
| subaru | impreza awd | 26.00000 |
| volkswagen | gti | 27.40000 |
| volkswagen | jetta | 29.11111 |
| volkswagen | new beetle | 32.83333 |
| volkswagen | passat | 27.57143 |
You would have made a double loop with condition. With 10 000 000 rows, 100000 groups you can't. Here it is efficient and concise. Alternative :
library(dplyr) # more or less equal perf
library(stringr) # to deal efficiently with strings
dose <- c("20 g/kg", "30g/kg lkd","15000 mg/kg")
dose
- '20 g/kg'
- '30g/kg lkd'
- '15000 mg/kg'
dose_corr <- data.frame( unit1 = str_extract(dose,"[a-z]+(?=/)"),
unit2 = str_extract(dose,"(?<=/)[a-z]+"),
value = str_extract(dose,"[0-9]+"))
dose_corr
| unit1 | unit2 | value |
|---|---|---|
| g | kg | 20 |
| g | kg | 30 |
| mg | kg | 15000 |
library(lmerTest)
library(lme4)
regressions linéaires, non linéaires, multiniveau, lineaire généralisé etc
a1_mean = 3
b1_mean = 0
curves <- data.table( curve = rep(letters[1:5],each = 10),
time = rep(1:10,5),
a1 = rep(rnorm(5,a1_mean,1),each = 10),
b1 = rep(rnorm(5,b1_mean,1),each = 10))
curves[, y_data := a1*time + b1 + rnorm(.N,0,2), by = curve]
curves[, y_real := a1*time + b1 , by = curve]
Create 5 curves with slope normaly distrib around a1_mean and intercept around b1_mean, with a bit of noise.
p <- ggplot(data = curves )+
geom_point(aes(time,y_data,color = curve),size = 3)+
geom_line(aes(time,y_real,color= curve),linetype = "dashed")+
theme_light()
p
fit <- lmer(y_data ~ time + (1 + time|curve) , data = curves)
summary(fit)
Linear mixed model fit by REML t-tests use Satterthwaite approximations to
degrees of freedom [lmerMod]
Formula: y_data ~ time + (1 + time | curve)
Data: curves
REML criterion at convergence: 242
Scaled residuals:
Min 1Q Median 3Q Max
-1.96349 -0.76208 -0.08607 0.71772 1.87458
Random effects:
Groups Name Variance Std.Dev. Corr
curve (Intercept) 0.1645 0.4055
time 1.1770 1.0849 -1.00
Residual 5.1073 2.2599
Number of obs: 50, groups: curve, 5
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 0.2499 0.7138 19.4780 0.350 0.73002
time 3.2979 0.4978 4.0060 6.625 0.00268 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr)
time -0.439
curves[,y_fit := predict(fit, newdata = curves)]
p + geom_line(aes(time,y_fit,color= curve),linetype = "solid")
Some others¶
- librudate for date handling
- rshiny to generate java/html interactives pages with buttons and graphs (amazing)
- reporters to generate table directly in word
- parallel to do parallel calculation
now real physicist examples:¶
- the join experiment I did with PSI in 2014.
- one hour in R , weeks during my post doc and matlab.
The experiment:
three main devices :
- arduino, say if laser is on or off
- grimm for particles sizes
- AMS, for composition (and size)
library(ggplot2)
library(data.table)
library(stringr)
library(lubridate)
setwd("C:/Users/dmongin/Documents/presentation_R_pourlesnuls/PSI")
list.files(pattern = ".txt$")
- 'arduino_tot.txt'
- 'grimm_tot.txt'
- 'grimm_tot_M.txt'
- 'testo_tot.txt'
- 'timeserie_ams4.txt'
arduino <- fread("arduino_tot.txt")
head(arduino)
| V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | V10 |
|---|---|---|---|---|---|---|---|---|---|
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.35 | 45.6 | 0 |
Here a stupid person didn't put any name (and its me), and didn't use any formating for the date. Here is the date formating
arduino[,Date := ymd_hms(paste0(V1,".",V2,".",V3," ",V4,":",V5,":",V6))] # make a string with all year month day and HMS
head(arduino)
| V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | V10 | Date |
|---|---|---|---|---|---|---|---|---|---|---|
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 | 2014-06-17 16:59:28 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 | 2014-06-17 16:59:28 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 | 2014-06-17 16:59:28 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 | 2014-06-17 16:59:28 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.00 | 45.6 | 0 | 2014-06-17 16:59:28 |
| 2014 | 6 | 17 | 16 | 59 | 28 | 61168 | 0.35 | 45.6 | 0 | 2014-06-17 16:59:28 |
arduino[,laser := V10]
arduino[,ozone := V9]
table(arduino[,.N,by = Date]$N) # table of line number per date (i.e. per second)
1 2 3 8 11 29 45
86003 165 1 1 2 1 1
Multiple lines with the same exact date !! Only one measure per second and variable of interest:
arduino[,N := .N,by = Date] # N is the number of line per Date
arduino_clean <- arduino[N == 1,.(Date,laser,ozone)] # subselection
head(arduino_clean)
| Date | laser | ozone |
|---|---|---|
| 2014-06-17 16:59:29 | 0 | 49.2 |
| 2014-06-17 16:59:30 | 0 | 49.2 |
| 2014-06-17 16:59:31 | 0 | 49.2 |
| 2014-06-17 16:59:32 | 0 | 49.2 |
| 2014-06-17 16:59:33 | 0 | 49.2 |
| 2014-06-17 16:59:34 | 0 | 49.2 |
ggplot(data = arduino_clean,aes(Date,ozone))+ # define data, x and y
geom_line()+ # curve type
geom_ribbon(aes(Date,ymin = 0,ymax = laser*500),fill = "red", alpha = 0.1)+ # make area trasparent
theme_light()+ # choose theme
facet_wrap(~date(Date),scales = "free") # one plot per day
testo <- fread("testo_tot.txt") # read
testo[,Date := ymd_hms(paste0(V1,".",V2,".",V3," ",V4,":",V5,":",V6))]# define the date
testo[,temp := V9] # define temp column
testo[,humidity := V8]
testo_clean <- testo[,.(Date,temp,humidity)] # subselect the columns of interest
head(testo_clean)
| Date | temp | humidity |
|---|---|---|
| 2014-06-17 16:59:35 | 24.802 | 40.739 |
| 2014-06-17 16:59:40 | 24.840 | 38.029 |
| 2014-06-17 16:59:45 | 24.819 | 38.965 |
| 2014-06-17 16:59:50 | 24.271 | 40.590 |
| 2014-06-17 16:59:55 | 24.440 | 40.947 |
| 2014-06-17 17:00:00 | 24.594 | 41.591 |
plouf <- merge(testo,arduino_clean,all.x = T,on = "Date") # merge arduino and testo
ggplot(data = plouf)+ # plot
geom_line(aes(Date,temp,color = "temperature"))+
geom_line(aes(Date,humidity,color = "humidity"))+
geom_ribbon(aes(Date,ymin = 0,ymax = laser*100,fill = "laser"), alpha = 0.2)+
theme_light()+ facet_wrap(~date(Date),scales = "free")
summary(lm(ozone ~ laser + humidity,data = plouf))
Call:
lm(formula = ozone ~ laser + humidity, data = plouf)
Residuals:
Min 1Q Median 3Q Max
-103.64 -23.30 -4.18 13.89 456.08
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 29.18843 1.09169 26.74 <2e-16 ***
laser 67.01037 0.52723 127.10 <2e-16 ***
humidity 0.15619 0.01437 10.87 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 56.69 on 53319 degrees of freedom
(700 observations deleted due to missingness)
Multiple R-squared: 0.2326, Adjusted R-squared: 0.2326
F-statistic: 8080 on 2 and 53319 DF, p-value: < 2.2e-16
summary(lm(humidity ~ laser + temp,data = plouf))
Call:
lm(formula = humidity ~ laser + temp, data = plouf)
Residuals:
Min 1Q Median 3Q Max
-24.3458 -2.7637 0.3438 3.4444 27.4333
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 121.593124 0.085689 1419.00 <2e-16 ***
laser -1.377861 0.056112 -24.56 <2e-16 ***
temp -2.575838 0.004224 -609.87 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 6.051 on 53319 degrees of freedom
(700 observations deleted due to missingness)
Multiple R-squared: 0.876, Adjusted R-squared: 0.876
F-statistic: 1.883e+05 on 2 and 53319 DF, p-value: < 2.2e-16
grim <- fread("grimm_tot.txt") # read file
setnames(grim,"[d&t31.12.2035 11:50:55]","date") # change weird name
grim[,Date := dmy_hms(date)] # make date
grim[,N := .N,by = Date] # count the number of line per date
grim <- grim[N == 1] # select just one line per s
head(grim)
| date | Counts-1320 [1/l] | Diameter-1320 [nm] | 0.265 µm | 0.290 µm | 0.325 µm | 0.375 µm | 0.425 µm | 0.475 µm | 0.540 µm | ... | 11.250 µm | 13.750 µm | 16.250 µm | 18.750 µm | 22.500 µm | 27.500 µm | 31.000 µm | 34.000 µm | Date | N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17.06.2014 17:01:10 | 100 | 0 | 2075 | 700 | 325 | 275 | 50 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2014-06-17 17:01:10 | 1 |
| 17.06.2014 17:01:30 | 100 | 0 | 2325 | 1175 | 800 | 325 | 100 | 50 | 25 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2014-06-17 17:01:30 | 1 |
| 17.06.2014 17:01:40 | 100 | 0 | 2650 | 1000 | 675 | 350 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2014-06-17 17:01:40 | 1 |
| 17.06.2014 17:01:50 | 100 | 0 | 2025 | 1075 | 625 | 400 | 150 | 25 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2014-06-17 17:01:50 | 1 |
| 17.06.2014 17:02:10 | 100 | 0 | 2000 | 1350 | 675 | 300 | 100 | 0 | 25 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2014-06-17 17:02:10 | 1 |
| 17.06.2014 17:02:20 | 100 | 0 | 2375 | 1425 | 425 | 375 | 250 | 0 | 25 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2014-06-17 17:02:20 | 1 |
I switch to long format instead of broad:
grim_clean <- melt(grim,measure.vars = patterns("m$"),variable.name = "size",value.name = "concentration")
head(grim_clean)
| date | Counts-1320 [1/l] | Diameter-1320 [nm] | Date | N | size | concentration |
|---|---|---|---|---|---|---|
| 17.06.2014 17:01:10 | 100 | 0 | 2014-06-17 17:01:10 | 1 | 0.265 µm | 2075 |
| 17.06.2014 17:01:30 | 100 | 0 | 2014-06-17 17:01:30 | 1 | 0.265 µm | 2325 |
| 17.06.2014 17:01:40 | 100 | 0 | 2014-06-17 17:01:40 | 1 | 0.265 µm | 2650 |
| 17.06.2014 17:01:50 | 100 | 0 | 2014-06-17 17:01:50 | 1 | 0.265 µm | 2025 |
| 17.06.2014 17:02:10 | 100 | 0 | 2014-06-17 17:02:10 | 1 | 0.265 µm | 2000 |
| 17.06.2014 17:02:20 | 100 | 0 | 2014-06-17 17:02:20 | 1 | 0.265 µm | 2375 |
I change the size to numbers
grim_clean[,size := as.numeric(str_extract(size,"[0-9]+\\.[0-9]+"))] # extract just numbers of size
grim_clean <- grim_clean[,.(Date,size,concentration)]
head(grim_clean)
| Date | size | concentration |
|---|---|---|
| 2014-06-17 17:01:10 | 0.265 | 2075 |
| 2014-06-17 17:01:30 | 0.265 | 2325 |
| 2014-06-17 17:01:40 | 0.265 | 2650 |
| 2014-06-17 17:01:50 | 0.265 | 2025 |
| 2014-06-17 17:02:10 | 0.265 | 2000 |
| 2014-06-17 17:02:20 | 0.265 | 2375 |
Example : plot the mean concentration per hour, for each day of the experiment:
moy_grim <- grim_clean[,.(concentration_m = mean(concentration)),by = .(size,hour(Date),date(Date))]
head(moy_grim)
| size | hour | date | concentration_m |
|---|---|---|---|
| 0.265 | 17 | 2014-06-17 | 43433.83 |
| 0.265 | 18 | 2014-06-17 | 28027.25 |
| 0.265 | 19 | 2014-06-17 | 20778.73 |
| 0.265 | 17 | 2014-06-18 | 69960.71 |
| 0.265 | 18 | 2014-06-18 | 68438.87 |
| 0.265 | 19 | 2014-06-18 | 74782.11 |
ggplot(data = moy_grim,aes(as.numeric(size),concentration_m))+
geom_line(aes(color = as.factor(hour),group = hour))+
facet_wrap(~date,scales = "free")+ theme_light() +
scale_y_log10() +
labs(x = "size um", y = "concentration", color = "day hour")
Warning message: "Transformation introduced infinite values in continuous y-axis"
AMS <- fread("timeserie_ams4.txt")
AMS[,Date := dmy_hms(time_stamp)]
head(AMS)
| time_stamp | Tot | Org | SO4 | NO3 | NH4 | Chl | Org43 | Org44 | Date |
|---|---|---|---|---|---|---|---|---|---|
| 18:06:2014 22:17:49 | 6.81623 | 3.75423 | 1.33178 | 0.798375 | 0.931821 | 1.45099e-05 | 0.310066 | 0.651729 | 2014-06-18 22:17:49 |
| 18:06:2014 22:18:10 | 6.36363 | 3.51243 | 1.27318 | 0.726966 | 0.845720 | 5.33109e-03 | 0.302963 | 0.617536 | 2014-06-18 22:18:10 |
| 18:06:2014 22:18:31 | 6.18041 | 3.29982 | 1.34318 | 0.761861 | 0.769005 | 6.54514e-03 | 0.277555 | 0.589916 | 2014-06-18 22:18:31 |
| 18:06:2014 22:18:53 | 6.38510 | 3.49896 | 1.34846 | 0.704156 | 0.824175 | 9.34887e-03 | 0.283127 | 0.606004 | 2014-06-18 22:18:53 |
| 18:06:2014 22:19:14 | 6.45665 | 3.50046 | 1.39680 | 0.696631 | 0.857673 | 5.08134e-03 | 0.300025 | 0.637838 | 2014-06-18 22:19:14 |
| 18:06:2014 22:19:35 | 5.71712 | 3.15600 | 1.19864 | 0.570948 | 0.779798 | 1.17294e-02 | 0.281378 | 0.546123 | 2014-06-18 22:19:35 |
define three experiment (dont remember):
# define 3 experiment with the dates
exp1 <- ymd(c("2014-06-17","2014-06-18","2014-06-19"))
exp2 <- ymd(c("2014-06-23"))
exp3 <- ymd(c("2014-06-25","2014-06-26"))
# for the date in each group, define new variable exp
grim_clean[date(Date) %in% exp1,exp := "exp1"]
grim_clean[date(Date) %in% exp2,exp := "exp2"]
grim_clean[date(Date) %in% exp3,exp := "exp3"]
AMS[date(Date) %in% exp1,exp := "exp1"]
AMS[date(Date) %in% exp2,exp := "exp2"]
AMS[date(Date) %in% exp3,exp := "exp3"]
I merge arduino results and AMS to have the laser
plouf <- merge(arduino_clean,AMS,all.y = T,by = "Date")
plouf <- plouf[!is.na(laser)]
head(plouf)
| Date | laser | ozone | time_stamp | Tot | Org | SO4 | NO3 | NH4 | Chl | Org43 | Org44 | exp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2014-06-18 22:17:49 | 0 | 35 | 18:06:2014 22:17:49 | 6.81623 | 3.75423 | 1.33178 | 0.798375 | 0.931821 | 1.45099e-05 | 0.310066 | 0.651729 | exp1 |
| 2014-06-18 22:18:10 | 0 | 34 | 18:06:2014 22:18:10 | 6.36363 | 3.51243 | 1.27318 | 0.726966 | 0.845720 | 5.33109e-03 | 0.302963 | 0.617536 | exp1 |
| 2014-06-18 22:18:31 | 0 | 33 | 18:06:2014 22:18:31 | 6.18041 | 3.29982 | 1.34318 | 0.761861 | 0.769005 | 6.54514e-03 | 0.277555 | 0.589916 | exp1 |
| 2014-06-18 22:18:53 | 0 | 33 | 18:06:2014 22:18:53 | 6.38510 | 3.49896 | 1.34846 | 0.704156 | 0.824175 | 9.34887e-03 | 0.283127 | 0.606004 | exp1 |
| 2014-06-18 22:19:14 | 0 | 33 | 18:06:2014 22:19:14 | 6.45665 | 3.50046 | 1.39680 | 0.696631 | 0.857673 | 5.08134e-03 | 0.300025 | 0.637838 | exp1 |
| 2014-06-18 22:19:35 | 0 | 33 | 18:06:2014 22:19:35 | 5.71712 | 3.15600 | 1.19864 | 0.570948 | 0.779798 | 1.17294e-02 | 0.281378 | 0.546123 | exp1 |
change to long format
AMS_long <- melt(plouf,measure.vars = c("Tot","Org","SO4","NO3","NH4","Chl","Org43","Org44"),value.name = "concentration",variable.name = "type")
head(AMS_long)
| Date | laser | ozone | time_stamp | exp | type | concentration |
|---|---|---|---|---|---|---|
| 2014-06-18 22:17:49 | 0 | 35 | 18:06:2014 22:17:49 | exp1 | Tot | 6.81623 |
| 2014-06-18 22:18:10 | 0 | 34 | 18:06:2014 22:18:10 | exp1 | Tot | 6.36363 |
| 2014-06-18 22:18:31 | 0 | 33 | 18:06:2014 22:18:31 | exp1 | Tot | 6.18041 |
| 2014-06-18 22:18:53 | 0 | 33 | 18:06:2014 22:18:53 | exp1 | Tot | 6.38510 |
| 2014-06-18 22:19:14 | 0 | 33 | 18:06:2014 22:19:14 | exp1 | Tot | 6.45665 |
| 2014-06-18 22:19:35 | 0 | 33 | 18:06:2014 22:19:35 | exp1 | Tot | 5.71712 |
# calculate the mean per aerosol type, laser state and experiment
result_AMS <- AMS_long[type != "Tot",.(concentration_m = mean(concentration)),by = .(laser,type,exp)]
# do the plot
ggplot(data = result_AMS,aes( as.factor(laser) ,concentration_m,fill = type))+
geom_col()+
facet_wrap(~ exp, scales = "free")+
labs( x = "laser", y = "concentration ppm", color = "type", fill = "type" ) + theme_light()
To visualize the time serie in time: not complicated either
ggplot(data = AMS_long[,laser_n := laser*max(concentration),by = exp],aes(Date,concentration))+
geom_ribbon(aes(Date,ymin = 0,ymax = laser_n),fill = "red", alpha = 0.1)+
geom_line(aes(colour = type))+ facet_wrap(~exp,scales = "free")+ theme_light()
Pitfalls¶
don't re-descover, use the libraries
use long format, grouping operation, and ggplot to make your life easy.
- Long format : each column a variable.
- Example grimm: All columns are particles concentration -> two columns, one concentration, the other size
spending time on data management makes life easier after. So:
- check if your unique values are unique
- always look out for missing values
- define the column type when importing data: make numeric variable numeric, avoid factors
Ressources¶
- http://r4ds.had.co.nz/ : R for Data science. Author is head of Rstudio, inventor of ggplot, dplyr, libridate, Rshiny etc. Ressource free online.
- stackoverflow ! Carefull:
- Check if the question is already asked
- Give a reproducible small example of your problem, so people can give solution(s)
- Be specific, show that you tried something
- If so, 20 minutes to have an answer max
- Rlunchs at unige ! http://use-r-carlvogt.github.io/prochains-lunchs/
Proof of warming in geneva in 20 lines of R¶
setwd("C:/Users/dmongin/Documents/presentation_R_pourlesnuls")
Geneva <- read.csv("test.txt",sep = ";",skip = 27)
head(Geneva)
Geneva <- setDT(Geneva)
| Year | Month | Temperature | Precipitation |
|---|---|---|---|
| 1864 | 1 | -4.2 | 13.6 |
| 1864 | 2 | -0.7 | 17.2 |
| 1864 | 3 | 5.3 | 32.7 |
| 1864 | 4 | 8.3 | 35.2 |
| 1864 | 5 | 13.5 | 68.9 |
| 1864 | 6 | 15.6 | 115.2 |
Geneva[,dec := Year %/% 10]
Geneva[,Temperature_dec := mean(Temperature),by = .(dec,Month)]
ggplot(data = Geneva,aes(Month,Temperature_dec))+
geom_line(aes(color = as.factor(dec*10)))+
theme_light()+
labs( color = "decennie", title = "mean per decennie", y = "Temperature", x = "Months" )
options(repr.plot.width=8, repr.plot.height=4.5)
library(zoo)
nyear <- 5
Geneva[,Temperature_rollmean := c(rollmean(Temperature,nyear,align = "center",fill = NA)), by = Month]
fit <- lmer(Temperature ~ I(Year/100) + (1+I(Year/100)|Month),data = Geneva)
Geneva[,Temperature_predict := predict(fit,newdata = Geneva)]
summary(fit)
Linear mixed model fit by REML t-tests use Satterthwaite approximations to
degrees of freedom [lmerMod]
Formula: Temperature ~ I(Year/100) + (1 + I(Year/100) | Month)
Data: Geneva
REML criterion at convergence: 7166
Scaled residuals:
Min 1Q Median 3Q Max
-5.5947 -0.6501 0.0148 0.6508 3.6801
Random effects:
Groups Name Variance Std.Dev. Corr
Month (Intercept) 89.59250 9.465
I(Year/100) 0.02251 0.150 -0.94
Residual 2.65071 1.628
Number of obs: 1855, groups: Month, 12
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) -15.96749 3.18911 11.02700 -5.007 0.000395 ***
I(Year/100) 1.31851 0.09514 11.01000 13.858 2.59e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr)
I(Year/100) -0.824
ggplot(data = Geneva)+
geom_point(aes(Year,Temperature_rollmean,color = as.factor(Month)))+
geom_line(aes(Year,Temperature_predict,color = as.factor(Month)))+
theme_light()+
labs( color = "Month", title = "Evolution", y = "Temperature", x = "Year" )
Warning message: "Removed 48 rows containing missing values (geom_point)."
calculation of gradient¶
taille <- 100
field <- data.table( x = rep(seq(-taille,taille,1),2*taille+1), y = rep(seq(-taille,taille,1),each = 2*taille+1))
field[,temp := exp(-(x^2 + y^2)/10000)]
p <- ggplot(data = field,aes(x,y)) +
geom_tile(aes(fill = temp))+
scale_fill_gradient(low = "green", high = "red")
p
gradx <- field[,.(gradx = c(NA,temp[3:.N],NA)-c(NA,temp[1:(.N-2)],NA), x = x),by = y] #gradient selon x
grady <- field[,.(grady = c(NA,temp[3:.N],NA)-c(NA,temp[1:(.N-2)],NA), y = y),by = x] # gradient selon y
grad <- merge(gradx,grady, by = c("x","y")) # merge des deux
# je prend 10 fleche par lignes, sinon y en a trop.
gradplot <- grad[x %in% seq(-taille,taille,round(taille/10)) & y %in% seq(-taille,taille,round(taille/10))]
# je renormalise le gradient pour que la longueur des fleches soit appreciable sur le plot
gradplot[,c("gradx","grady"):= .(gradx/max(gradx,na.rm = T)*10,grady/max(grady,na.rm = T)*10)]
p + geom_segment(data = gradplot,aes(x,y,xend =x+ gradx,yend = y+grady), arrow = arrow(length = unit(0.2,"cm")))
Warning message: "Removed 80 rows containing missing values (geom_segment)."