How to find someone in a data frame
An important technical problem when updating personal data
Context
During the COVID19 outbreak in Geneva, I worked at état de Genève as a data scientist. We set up the follow-up system of COVID19 patient in Geneva, based on redcap to allow to centrally store the data stemming from the different labs, from the hospitals, from the state. We used for all the data management and data handling, because I master it properly, because there are package allowing to interact directly with redcap via its API. But with the outbreak going on, we rapidly faced a problem: how to properly identify a person when you have different source of data? We needed to/
- detect potential duplicated person in our database
- update data that were already stored in redcap with new results, but coming from different sources, where the name and surname were sometime not exctly the same.
And actually both point are the same issue: how to identify a person.
I will detail here the different step of the solution I implemented, beginning with the second point: how to detect duplicated person in a data frame. It involves a bit of regex, some fuzzy matching concepts, some clustering analysis, a bit of graph analysis, and some nice logical tricks.
The libraries:
library(data.table)
library(dplyr)
library(igraph)
library(stringdist)
library(qualV)
# for this page:
library(DT)
library(viridis)I use data.table, but little is required. I strongly encourage using this package when dealing with data (see here for a nice discussion about it).
You will find nice explanation and examples in the vignettes of this package, but here are the basics of what I will use:
dt <- data.table(a = sample(1:10,10,replace = T),ID = sample(1:3,10,replace = T))
dt## a ID
## 1: 9 1
## 2: 6 1
## 3: 10 1
## 4: 1 2
## 5: 6 3
## 6: 9 2
## 7: 7 2
## 8: 8 1
## 9: 7 2
## 10: 4 2To assigne a new column:
dt[,b := a + 5]To subset:
dt[a == 6]## a ID b
## 1: 6 1 11
## 2: 6 3 11To do grouping operations:
dt[,mean(a),by = ID]## ID V1
## 1: 1 8.25
## 2: 2 5.60
## 3: 3 6.00I use .GRP which is a help returning the grouping index:
dt[,.GRP,by = ID]## ID GRP
## 1: 1 1
## 2: 2 2
## 3: 3 3dt[,.GRP,by = .(ID,a)]## ID a GRP
## 1: 1 9 1
## 2: 1 6 2
## 3: 1 10 3
## 4: 2 1 4
## 5: 3 6 5
## 6: 2 9 6
## 7: 2 7 7
## 8: 1 8 8
## 9: 2 4 9That’s it.
Let’s find them
I will detail the steps on a simple example:
df <- data.table(surname = c("Mongin","mongin","smith","De la cruz","de-la-cruz","smith","mongin","SMITH","smith","Smith","Cruz"),
name = c("denis","denis diego","paul","sofia","sophia, Elèna","Paul","dénis","Paul ","Etienne","Paule","sofia"),
birthdate = c("29-08-1986","29-08-1986","02-06-1945","02-03-2000","02-03-2000","20-06-1945","29-07-1986","02-06-1945","02-06-1945","02-06-1945","02-03-2000"))
Here we have 4 people, and we want to identify them properly. The name sometime changes, because some data store all the names when other just the first, they are some typos on the birthdates, etc. Real life data.
Normalize the names and surnames
The first thing to do is to try to get the name and surname on the same format: removing white spaces, punctuation marks, accents, and putting everything lower case:
clean_name <- function(name) {
name_clean <- name %>%
as.character() %>%
tolower(.) %>%
chartr("âäëéèêóôöüûçîï", "aaeeeeooouucii", .) %>%
gsub("\\bla\\b|\\bde\\b|\\bda\\b|\\bdu\\b|\\bl'\\b", "", .) %>%
gsub("[ ]+|[-]|[,]|[\\.]", "", .)
return(name_clean)
}
clean_name(df$name)## [1] "denis" "denisdiego" "paul" "sofia" "sophiaelena"
## [6] "paul" "denis" "paul" "etienne" "paule"
## [11] "sofia"cat("\n\n")clean_name("testéDetru -bizarùe.")## [1] "testedetrubizarùe"Here:
chartrchange one character for another. I use it to remove the accents.- In regex:
-
\\bindicates a word boudary: I remove allde,duand other small word of composed surnames.|is for or
It is acually a good start, but it is not enough: If I clean names and surnames in the data:
df[,name_clean := clean_name(name)]
df[,surname_clean := clean_name(surname)]and associate a person index for all person with same cleaned name, surname and birthdate:
df[,person := .GRP,by = .(name_clean,surname_clean,birthdate)]I recognize some of these persons, but I miss most of them:
fuzzy matching
The comparison function
So, I need to let a bit of variation in my character recognition. There are quite a lot of possibility with the library stringdist, but most of them did not solve the problem I had with the name, which is the following: denis and denisdiego should be the same, but denis and yanis should not. If I try
stringdist("denis","denisdiego")## [1] 5stringdist("denis","yanis")## [1] 2The distance is greater for the first case, because I need to add 5 characters to go from denis to denisdiego, while I only need to change two characters to switch from denis to yanis.
My solution uses the qualV package. Here is the first function I did:
library(qualV)
comp_names <- function(a, b) {
a <- clean_name(a) # cleaning"
b <- clean_name(b)
av <- strsplit(a, '')[[1]] # splitting all characters
bv <- strsplit(b, '')[[1]]
match_str <- paste(qualV::LCS(av, bv)$LCS, collapse='')
pc <- round(nchar(match_str)/(min(nchar(a), nchar(b)))*100)
return(pc)
}I clean the two names a and b, cut it in sequence of character, fin the longest common subsequence, and express it as a percentage of the smallest name. It outputs a similarity on a 100 scale:
comp_names("denis","denisdiego")## [1] 100denis and denisdiego are totally similar, while
comp_names("denis","yanis")## [1] 60denis and yanis are not so much the same.
Matrix of distances and clustering
I need to calculate the distance between all the entries of a vector, i.e. I need to compute a distance matrix. The distance between two names Tis just 100 - similarity I calculate before. Here is the function to generate the distance matrix:
comp_names_matrix <- function(vec) {
mat <- sapply(vec, function(x) {
100 - as.numeric(Vectorize(comp_names)(x, vec))
})
rownames(mat) <- vec
return(mat)
}comp_names_matrix(df$name)## denis denis diego paul sofia sophia, Elèna Paul dénis Paul
## denis 0 0 100 80 60 100 0 100
## denis diego 0 0 100 60 70 100 0 100
## paul 100 100 0 75 25 0 100 0
## sofia 80 60 75 0 20 75 80 75
## sophia, Elèna 60 70 25 20 0 25 60 25
## Paul 100 100 0 75 25 0 100 0
## dénis 0 0 100 80 60 100 0 100
## Paul 100 100 0 75 25 0 100 0
## Etienne 60 57 100 80 57 100 60 100
## Paule 80 80 0 80 20 0 80 0
## sofia 80 60 75 0 20 75 80 75
## Etienne Paule sofia
## denis 60 80 80
## denis diego 57 80 60
## paul 100 0 75
## sofia 80 80 0
## sophia, Elèna 57 20 20
## Paul 100 0 75
## dénis 60 80 80
## Paul 100 0 75
## Etienne 0 80 80
## Paule 80 0 80
## sofia 80 80 0Now I can use the basic tools of clustering to see if I can identify the similar names:
df$name %>%
comp_names_matrix(.) %>%
stats::as.dist(.) %>%
stats::hclust(.) %>%
plot 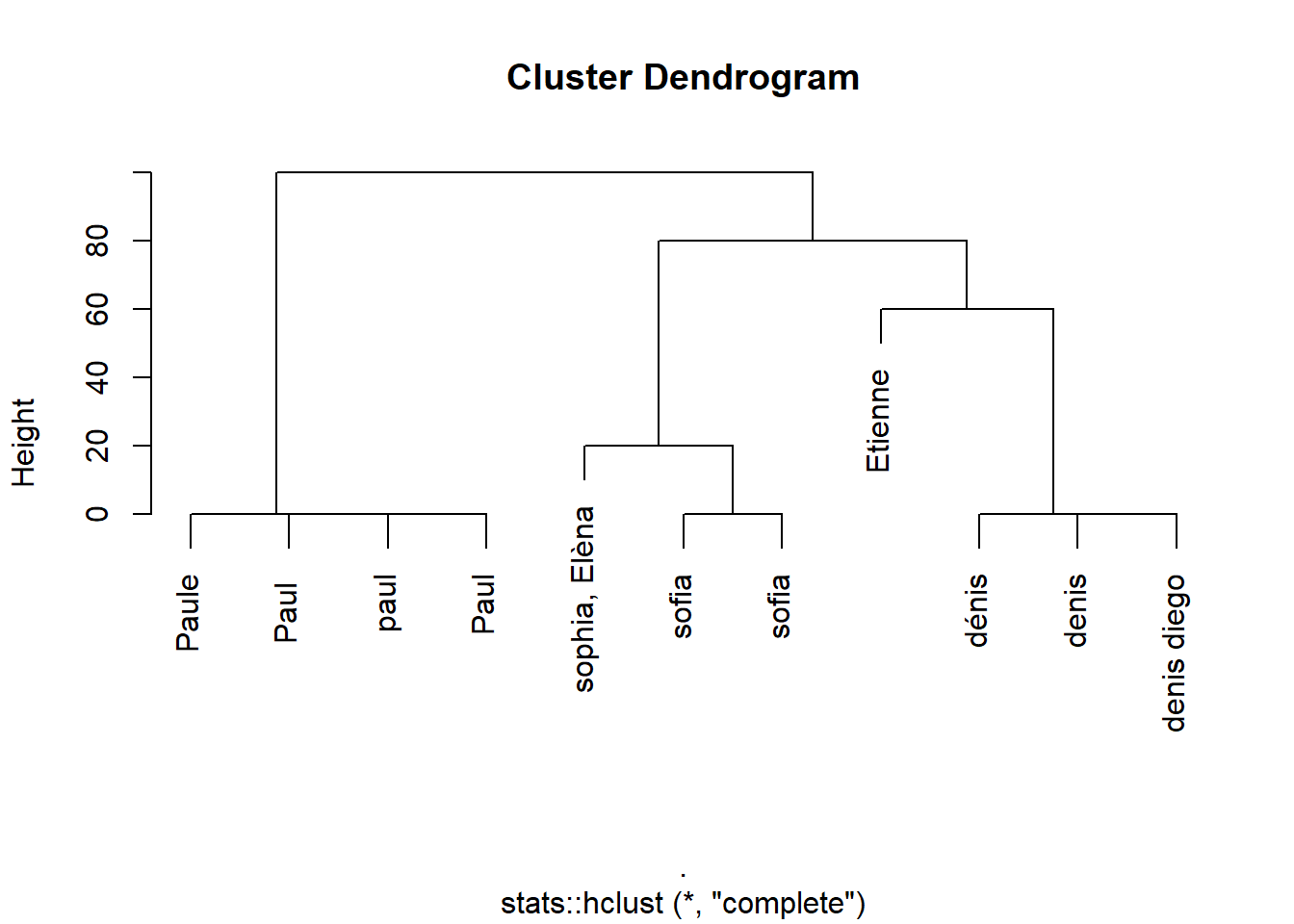
Here it seems that my distance allow me to detect the similar person in a proper way. I need to find a threshold to produce an index indicating the names that are the same. I see in the dendrogram above that it lies between 20 and 40. The function to use is stats::cutree(h = thres). I write the following, that use the name comparison function described above and detect clusters of names:
compare_tree <- function(vec, thres = 30) {
if(length(vec)>1){
output <- vec %>%
comp_names_matrix(.) %>%
stats::as.dist(.) %>%
stats::hclust(.) %>% # cluster
stats::cutree(h = thres)}else{output <- as.integer(1)}
return(output)
}
compare_tree(df$name,30)## denis denis diego paul sofia sophia, Elèna
## 1 1 2 3 3
## Paul dénis Paul Etienne Paule
## 2 1 2 4 2
## sofia
## 3It recognize the 4 names.
I can now apply this function on the names, surnames and birthdates too.
df[,name_ind := compare_tree(name,30)]
df[,surname_ind := compare_tree(surname,30)]
df[,birthdate_ind := compare_tree(birthdate,20)]I define a person index for the common detected name, surname and birthdate:
df[,person := .GRP,by = .(birthdate_ind,surname_ind,name_ind)]It worked. It is flexible enough to allow small mistakes in birthdate and can handle the fact that sometime there is just one name, sometime there are the 3 names of that same person.
If I do that on my entire COVID19 patient table, my computer turn into that:

I have around 70000 patients, which lead to 4.910^{9} distances to calculate. My solution is nice but cannot be used in a real life situation. I therefore need to constrain the degrees of freedom.
Less degrees of freedom
To avoid calculation of immense distance matrix, I can do grouping operation. What I came up with is to calculate the similarity of the names for person having identical cleaned surname and birthdate:
df[,idx1 := paste0("1_",.GRP,"_",compare_tree(name,30)),
by = .(birthdate,surname_clean)]The index is constructed as follow: it starts with 1, then it has the grouping index produced by .GRP, and then the index indicating similar names.
This gives me a first index: person with same birthdate and surname, and similar names. I do the same for names and birthdate:
df[,idx2 := paste0("2_",.GRP,"_",compare_tree(surname,30)),
by = .(name_clean,birthdate)]and the last one:
df[,idx3 := paste0("3_",.GRP,"_",compare_tree(birthdate,20)),
by = .(name_clean,surname_clean)]You see that line 1 and 2 have the same idx1 index: same surname, same birthdate, and similar name detected by my function. You can see also that line 1 and 7 have the same idx3 index: same name, surname, and similar birthdates.
I need to somehow reconnect these indices to merge them into one, indicating that the lines 1, 2 and 7 are the same person. The answer came from stack: we need to do graph analysis and identify clusters. We can define the lines as nodes, connected by the 3 indices. Nodes connected are the same person. To do that, I use the igraph library:
df[, rn := .I]
DT <- rbindlist(list(
df[, .(s=idx1, e=idx2, rn)],
df[, .(s=idx1, e=idx3, rn)],
df[, .(s=idx2, e=idx3, rn)]))df[, rn := .I] just create the row number rn (.I is a shortcut for the row number in data.table, really useful in some complex grouping operation). DT is the list of edges and vertices. Here is the graph:
g <- graph_from_data_frame(DT, directed=FALSE)
print(g)## IGRAPH ea9b148 UN-- 22 33 --
## + attr: name (v/c), rn (e/n)
## + edges from ea9b148 (vertex names):
## [1] 1_1_1--2_1_1 1_1_1--2_2_1 1_2_1--2_3_1 1_3_1--2_4_1 1_3_1--2_5_1
## [6] 1_4_1--2_6_1 1_5_1--2_7_1 1_2_1--2_3_1 1_2_2--2_8_1 1_2_1--2_9_1
## [11] 1_3_1--2_4_1 1_1_1--3_1_1 1_1_1--3_2_1 1_2_1--3_3_1 1_3_1--3_4_1
## [16] 1_3_1--3_5_1 1_4_1--3_3_1 1_5_1--3_1_1 1_2_1--3_3_1 1_2_2--3_6_1
## [21] 1_2_1--3_7_1 1_3_1--3_4_1 2_1_1--3_1_1 2_2_1--3_2_1 2_3_1--3_3_1
## [26] 2_4_1--3_4_1 2_5_1--3_5_1 2_6_1--3_3_1 2_7_1--3_1_1 2_3_1--3_3_1
## [31] 2_8_1--3_6_1 2_9_1--3_7_1 2_4_1--3_4_1I then detect clusters:
cl <- clusters(g)$membership
cl## 1_1_1 1_2_1 1_3_1 1_4_1 1_5_1 1_2_2 2_1_1 2_2_1 2_3_1 2_4_1 2_5_1 2_6_1 2_7_1
## 1 2 3 2 1 4 1 1 2 3 3 2 1
## 2_8_1 2_9_1 3_1_1 3_2_1 3_3_1 3_4_1 3_5_1 3_6_1 3_7_1
## 4 2 1 1 2 3 3 4 2DT[, g := cl[s]]And I merge back the found index in my initial data table, merging on the row number:
df[unique(DT, by="rn"), on=.(rn), person := g]That’s it !
I put everything in a function:
Finding someone in a dataframe
Now, how can I find someone in my example data.frame ? Let us suppose I want to find these two persons in my example data frame:
tofind <- data.table(surname = c("mongin","Cruz, de la"),name = c("denis","Sophia"),birthdate = c("29-09-1986","02-03-2000"))
tofind## surname name birthdate
## 1: mongin denis 29-09-1986
## 2: Cruz, de la Sophia 02-03-2000Well, the nice thing is that I can use the exact same function to do so. I first create a variable to indicate the lines and name of each data frame:
tofind[,rn_df := .I]
df[,rn_df := .I]
tofind[,dfframe := "tosearch"]
df[,dfframe := "database"]and I bind the two together
dftot <- rbind(tofind,df[,.(name,surname,birthdate,rn_df,dfframe)])Looking for someone is the same as detecting the duplicates in the binded data frame ! If I use my function to detect the person on dftot:
res <- detect_duplicate(dftot)I can easily see which line comes from the data frame with people I wanted to find:
I can now easily extract the correspondence between the lines of each data frame:
dftot[,expand.grid(rn_tofind = rn_df[dfframe == "tosearch"],
rn_database = rn_df[dfframe == "database"]),
by = person]## person rn_tofind rn_database
## 1: 1 1 1
## 2: 1 1 2
## 3: 1 1 7
## 4: 2 2 4
## 5: 2 2 5
## 6: 2 2 11The line 1 of my data frame with the people I was searching mongin, denis, 29-09-1986, 1, tosearch is found on line 1, 2 and 7 of my example data base, while the second person Cruz, de la, Sophia, 02-03-2000, 2, tosearch is on line 4, 5 and 11 of my data base.
Conclusion
The function here is a simplified version of the one we use, but it contains all the core function that makes it efficiently works. It became a central part of the COVID data handling in Geneva, as we use it to update all the data of our data base directly from R: test results, deaths, hospitalizations, contacts etc.